The paper in Nature Communications is here: go.nature.com/2xay9Ui
Genome engineering has been surprisingly harnessed through the development of the CRISPR/CRISPR-associated protein (Cas) system [1, 2], and researchers have expected that CRISPR from Prevotella and Francisella 1 (Cpf1) would be a wonderful alternative tool for genome editing. When it has been reported that CRISPR-Cas9 recognizes a 20-nt target sequence via crRNA-target strand base pairing, we thought that 20-nt is a enough length for a specific recognition. We have already seen the similar case in ZFN and TALEN. That's why we are somehow puzzled when CRISPR-Cpf1 has been reported to use crRNA carrying 23- or 24-nt target complementary sequence [3]. It may be that the extended length is to make sure that the target recognition is more specific. However, several structural analyses [4, 5] indicated that it's not that way. The 3'-terminal 3-nt crRNA sequence is so flexible that the region is not identified in the X-ray crystallographic setting. We understood that this may mean the PAM-proximal 3-nt sequence in crRNA does not participate in the target binding. Rather, it may play an auxilliary role. This question urged us to investigate the role of the extra 3-nt sequences of crRNA in CRISPR-Cpf1 system.
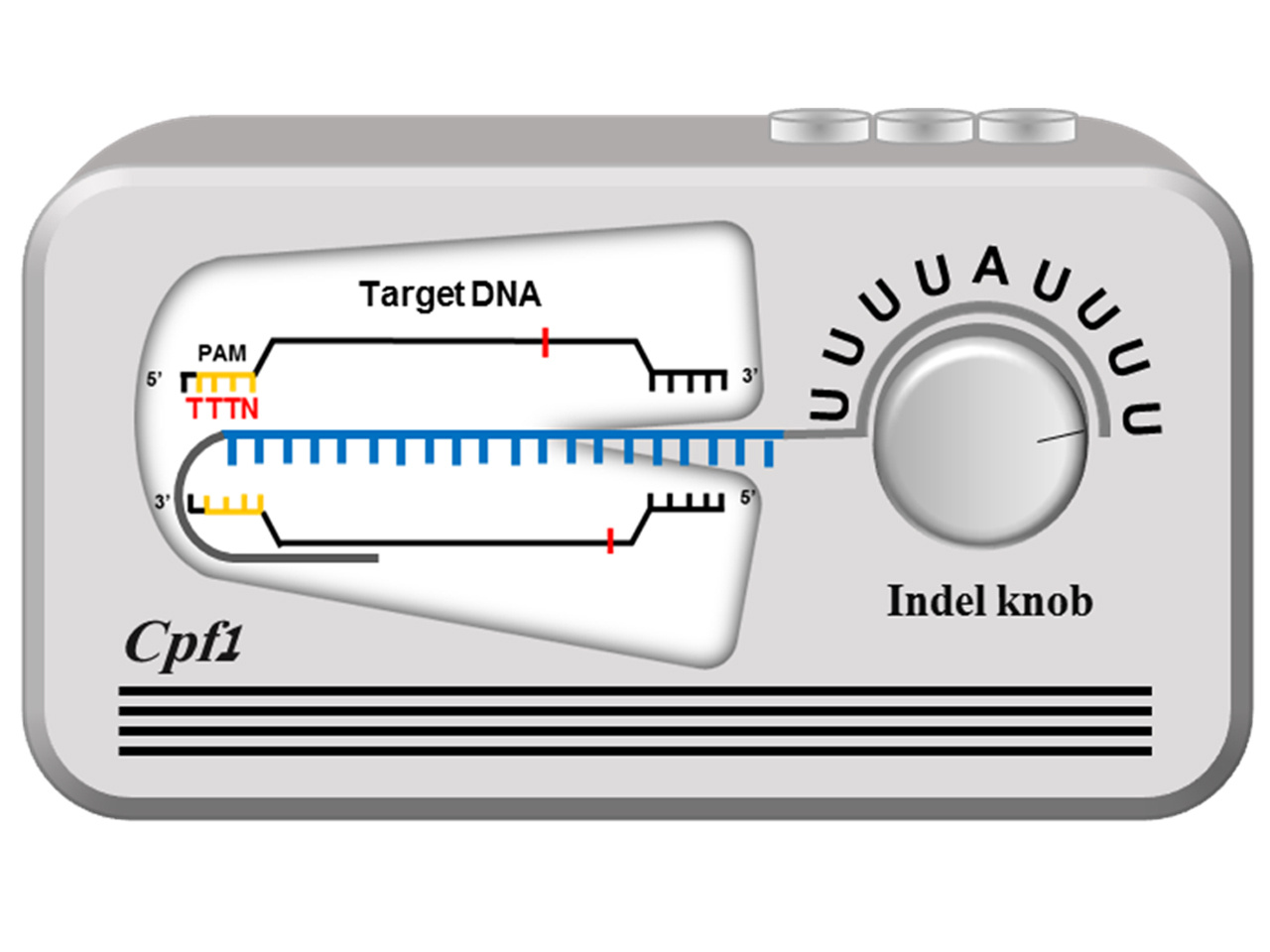
The first thing we did was to change the PAM proximal 3-nt sequence into non-target sequences and to compare those with a target one. To our surprise, the triple U (U3) created an improved indel efficiency in HEK-293T cells (Fig. 1a in the original paper). This important finding made us test the effect of 3'-terminal sequences on the indel activity of Cpf1. The rigorous in vitro and in vivo experiment revealed that the presence of uridinylates up to ~8 nt imposed the highest activity on the crRNA-Cpf1 complex. When crRNA is transcriptionally produced from the RNAIII-dependent U6 promoter, U5 or U6 act as a termination signal and thus the uridinylates can't be extended to 8-nt. That's why we incorporated A among the rich Us and completed the optimized configuration of the crRNA template that carrys a T4AT6 tail. I asked my student, Moon SB, who is the first author of this paper, to schematize our finding in a simple way, and she brilliantly completed the following sheme (Figure 1 below). Cpf1 system can be compared to a radio set where the U-rich tail acts as a volume control knob. By controlling the length of U, one may regulate the Cpf1 activity. Wow, it's a tunable Cpf1 system.
CRISPR-Cpf1 shows multiple inherent advantages over CRISPR-Cas9 in that it has a smaller effector gene size, smaller guide RNA, low off-target activity, and complementary PAM sequences(A/T-rich). Astonishingly, it was recently reported in bioRxiv that a significant portion of humans have pre-existing adaptive immunity to Cas9 proteins. Nature responded to this report as "Researchers hoping to use a gene-editing technique to treat disease may have to seek alternative enzymes". All these facts emphasizes the potential advantages of CRISPR-Cpf1 system. Nonetheless, the adoption of Cpf1 has been much lower than expected. We evaluated that his startlingly low adotpion of Cpf1 originated from a lower effeciency and high target-to-target variations. Our large-scale validation demonstrated that the engineered Cpf1 system shows an indel activity comparable to Cas9. Given the innate multiple merits of Cpf1, there's no reason to prefer to use Cas9 instead of Cpf1 unless it's a PAM issue.

Figure 1. Scheme for a tunable, highly efficient CRISPR-Cpf1 with the uridinylate-rich 3'-tail.
References
1. Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823 (2013).
2. Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823-826 (2013).
3. Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759-771 (2015).
4. Dong, D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522-538 (2016).
5. Yamano, T. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949-962 (2016).





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in