Cell-free systems are widely used for fundamental molecular biology, for applications in genetic engineering, in metabolic engineering or in medical diagnostics and for a better understanding in the origin of life1,2,3. Cell-free systems present many advantages for synthetic biology through high-throughput characterization and prototyping of natural and synthetic circuits. Over the last decade, the cell-free community developed multiple protocols to simplify lysate preparation and maximize protein production in vitro. The lysate-based transcription-translation cell-free systems (TXTL) using E. coli can currently be prepared in a few days via chemical or physical disruption of the cellular membrane and mixed with an energy buffer.
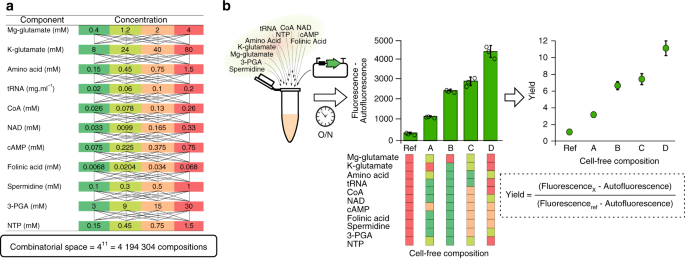
In Jean-Loup’s team, we based our work on E. coli cell-free system on the Sun et al. protocol4. We used sonication instead of bead beading to prepare our lysate but stick to the washing steps and buffer nicely described in the protocol. We noticed that a crucial step of the protocol requires an adjustment of two compounds concentration: Mg-glutamate and K-glutamate. Such lysate-specific adjustment was necessary after the preparation of every new batch of lysate to maximize protein production. Even with the same procedure, the homemade preparation of the cell-free mix led to variability of the protein production efficiency.
The above variabilities, makes predictions difficult and cell-free mix inefficient. We get an interest in eleven other compounds present in the buffer which interactions between each other are still unclear. Such interactions can change between lysate batch, as some compounds are already in the lysate, and affect the transcription and translation efficiencies. As our team is composed of a dry lab skilled in machine learning and a wet lab with experience in cell-free approach, it was natural to tackle this problem using the recent advance made in machine learning and in automation of biology. Machine learning approach avoids the requirement of multiple unverified assumptions and lead to impressive high quality predictions when properly trained. To start the project we wrote to Labcyte Inc. a company specialized in automation who Olivier had the chance to work with at the Imperial College London. They put us in contact with the Chemogenomic and Biological Screening core facility of Fabrice Agou at Institut Pasteur where they developed a "strong savoir-faire" in assay development as well as in high-throughput biological screening. Through this collaboration, we got help and advice on protein synthesis and screening technologies from Agou’s team and especially the expertize of Agnes to develop automation methods. The first step was to check how the acoustic Liquid handling (Echo Machine) managed each of the 11 compounds. We then worked on a set of controls (buffer compositions leading to a wide range of protein productions) to compare measurements between each iteration. When these preliminary steps were set up, Mathilde (dry lab) and Olivier (wet lab) worked on the iteration cycle:
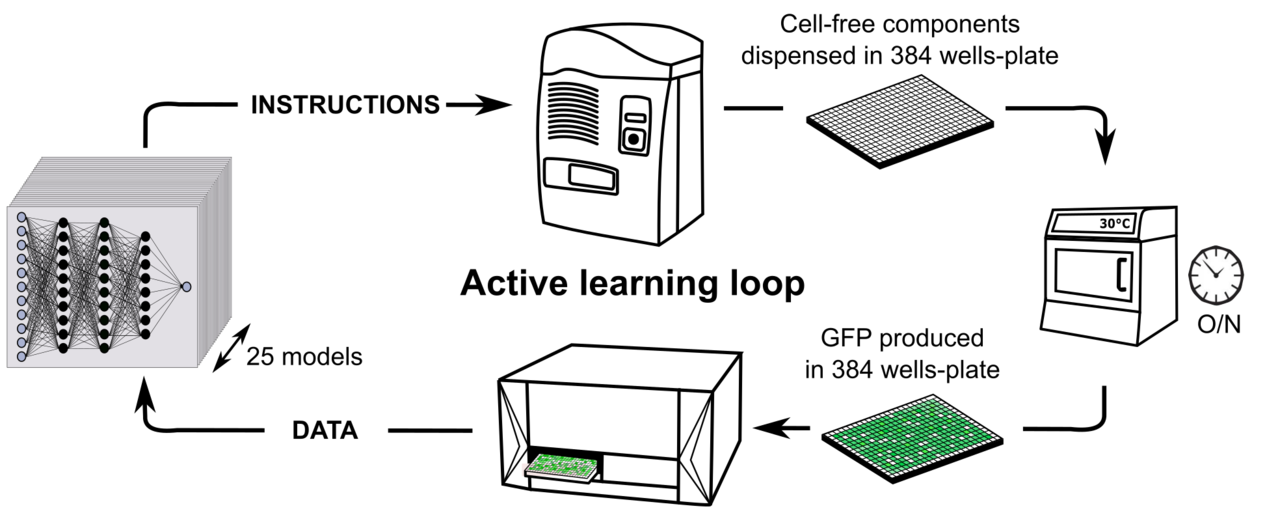
Provide robot instructions – Distribute compounds in 384 well plates – Incubation overnight – Fluorescence measurements – Prediction with Neural network Models.

A) Cell-free composition. We modified the concentration of eleven compounds in the cell free mix. B) Active learning loop for cell-free productivity predictions.
The stock of compounds and the lysate were prepared before distribution and the Echo Machine managed the equivalent of 4000 pipetting at every iteration. This process was a cycle of exchanges between the instructions provided by Mathilde and the measurements provide by Olivier.
We wanted to be sure to have enough lysate and plasmid DNA for the whole active learning cycles, so Amir and Olivier prepared a large batch of lysate with 12 liters of E. coli culture. Amir also provided a large stock of maxi prep DNA to measure ~10 000 reactions. The critical part was to provide sufficiently large dataset to train Mathilde’s neural network models and find the highest production level in a vast combinatorial space of more than 4 million possible buffer compositions. During the active learning, we found a maximum protein production after seven iterations (~2000 measurements) and obtain high-quality predictions with a correlation ~0.9 between predictions and measurements. The second objective was to extract a small subset of highly informative compositions for other users to train the models with data from their own lysates. We asked Paul and Angelo, two PhD students of the lab to prepare their own lysate with the same E. coli strain and protocol. Then, Mathilde checked if a subset of 20 buffer combinations was sufficient to predict protein production with other lysates. The trained models were very efficient with Paul and Angelo lysates and even when the transcription process was damaged in a lysate supplemented with an antibiotic. We used a little larger subset of 5 extra buffer combinations when using another strain of E. coli or when the translation process was damaged with a mix supplemented with an antibiotic.
During this study, we showed that the combination of machine learning, automation and in vitro protein synthesis is extremely efficient to predict genetic circuits output. Such interdisciplinary approach is of particular interest for the field of synthetic biology, which often stumbles upon difficulties to predict the outcome of genetic circuits with large combinatorial design. We hope that our results will extend the use of cell-free systems in wet lab and especially for molecular biologist by making easier the production of protein in vitro.
By Olivier Borkowski & Jean-Loup Faulon
Our full article is available here: https://www.nature.com/article...
References
1 Hodgman, C. E., & Jewett, M. C. Cell-free synthetic biology: Thinking outside the cell. Metab. Eng.14, 261-269 (2012).
2 Pardee, K., Green, A.A., Takahashi, M.K., Braff, D., Lambert, G., Lee, J.W., Ferrante, T., Ma, D., Donghia, N., Fan, M. , Daringer, N.M, Bosch I., Dudley D.M., O'Connor D.H., Gehrke L and Collins J.J. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell, 165(5), 1255-1266 (2016)
3 Laohakunakorn, N., Grasemann, L., Lavickova, B., Michielin, G., Shasein, A., Swank, Z. and Maerkl, S.J. Bottom-up construction of complex biomolecular systems with cell-free synthetic biology. Frontiers in Bioengineering and Biotechnology, 8, p.213. (2020)
4 Sun, Z. Z., Yeung, E., Hayes, C. A., Noireaux, V., & Murray, R. M. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol.3, 387–397 (2014).


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in