I am still impressed by the scene depicted by Dr. Lin during my post-doctoral position interview when asking my future project in his lab. “Protein Post-translational modifications (PTMs) play central roles in a wide range of biological processes. However, it is still challenging and laborious to address the function of any particular modification by the traditional biochemical and genetic approaches. GCE technology allows site-specifically and co-translationally incorporation of authentic PTMs, enabling study the effect of these modifications directly by this gain-of-function mutation. Currently, very few PTMs can be efficiently incorporated in both prokaryotes and eukaryotes, due to the lack of fully orthogonal aaRS/tRNA pairs. The pyrrolysyl-tRNA synthetase/tRNA pair (PylRS/PylT) from Mm and Mb is the only available fully orthogonal system so far1. So, let’s focus on this issue and develop more fully orthogonal pairs to recognize PTMs of interest.”

Figure 1. The overview of our strategy and result highlights. (a) The schematic of chimera design, (b) Fully orthogonal pairs developed in this study, (c) UAAs incorporated with chPheRS/chPheT pair.
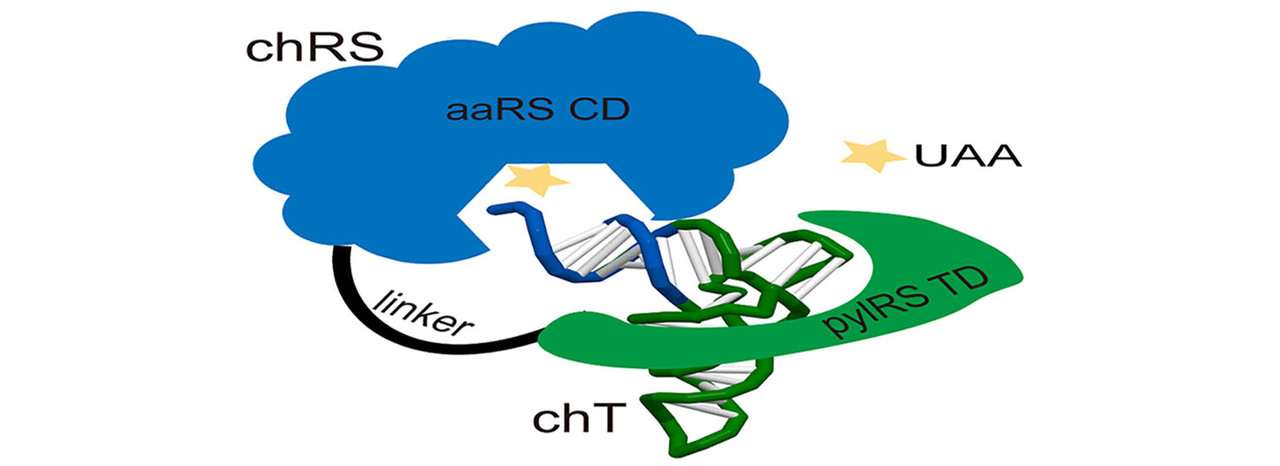
After joining his lab, we were discussing how to approach this issue very often. During one discussion, we realized that we can directly transplant the orthogonality of PylRS/PylT pair to canonical aaRS/tRNA pairs. we immediately surveyed the three-dimensional structure of all the canonical aaRS /tRNA pairs deposited in PDB and analyzed the spatial arrangement of their CD (catalytic domain) and TD (tRNA binding domain) combining biochemical data, finally chose HisRS/HisT pair from E.coli as starting point for the clear distinction between CD and TD. We firstly constructed a preliminary version of chimeric HisRS (chHisRS-1) by fusing PylRS TD at the N-terminus of HisRS CD. Then a series of chHisTs were constructed and estimated by GFP-amber suppression assay along with chHisRS-1. Luckily, a chHisT with intact acceptor stem from HisT achieved 35% efficiency compared with GFP-WT. We further demonstrated that PylRS TD-IPYE at N-terminus of fusion driven by strong promoter like oxb20 significantly increased efficiency. we subsequently proved chHisRS/chHisT pair was fully orthogonal and highly active for Histidine incorporation in E.coli and mammalian cells with in vivo and in vitro assay. With this successful experience in mind, we set out to apply it to other canonical synthetase/tRNA pairs. After hundreds of clones were built and tested, three additional fully orthogonal pairs-chPheRS/chPheT, chAlaRS/chAlaT and chSerRS/chSerT were created and could be sent to evolve to alter substrate specificity for UAAs incorporation.
We had noticed that double-Gly mutant of yeast PheRS had been utilized to incorporate several UAAs in E.coli2, so we next focused on chPheRS/chPheT pair to expand substrate scope. Double-Gly mutant of chPheRS (T467G/A507G, dubbed as chPheRS-1) was firstly constructed and found to incorporate 4-Azido-Phe effectively in E.coli and mammalian cells. Then three types of chPheRS mutations accommodating UAAs with distinct functionality were developed by selection and screen. ChPheRS-2 (Q356N/T467S/A507S) recognized L-Dopa and could be served as novel tool to study the function of Tyrosine hydroxylation in situ. ChPheRS-3 (E391D/T467G/A507G) and chPheRS-4 (F464V/T467G/A507G) preferred Trp analogs containing 6 and 7-cyano-Trp with unique fluorescence could be used to study protein localization and conformation change.
Currently, we have engineered four fully orthogonal chimera pairs, we are testing whether unrecognized PTMs can be incorporated by these systems through pocket engineering. At the same time, we are investigating the working mechanism of this chimera strategy and hoping to bring in more orthogonal pairs in the near future.
References:
1. Dumas A, Lercher L, Spicer CD, Davis BG. Designing logical codon reassignment - Expanding the chemistry in biology. Chemical Science 2015, 6(1): 50-69.
2. Kwon I, Tirrell DA. Site-specific incorporation of tryptophan analogues into recombinant proteins in bacterial cells. Journal of the American Chemical Society 2007, 129(34): 10431-10437.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in