As a synthetic biologist, you could be working with eukaryotes, prokaryotes, or even cell-free systems, but when it comes to controlling gene expression, we all face similar challenges. Often, it is difficult to ensure a gene is fully turned off, or sufficiently expressed when induced.
During my PhD, I wasn’t an exception to those issues.
Here is the story behind our paper (Greco et al. Nature Communications, 2021), where we found that by adding the right sort of complexity to our regulatory programmes, we could get gene expression under control and reap some added benefits.
“In the middle of difficulty lies opportunity” - Albert Einstein.
It all started when I was testing a set of inducible systems and fluctuations in gene expression when the system should have been ‘off’ kept jeopardising my results. Precise control is crucial when characterising parts in synthetic biology, but how could I collect high quality data if I couldn’t even get it to turn on or off correctly?
The natural world has always been an inspiration for scientists, who have learnt to mimic natural processes for centuries. Nature achieves control in many different ways and often by utilising regulation at many different levels. For example, the Venus flytrap controls the closing of its traps via trigger hairs that detect a crawling insect. To make sure the traps don’t accidentally close, the flytrap uses a mechanism whereby trap closure is only triggered if two mechanical stimulations occur (Scherzer et al. Nature Plants 5(7), 670-75, 2019). This more stringent control mechanism, although simple, builds in redundancy and helps ensure the plant makes the best use of its resources.
Mechanisms like this inspired me. I wondered if exploiting the multiple stages in gene expression might offer a solution. Maybe, I could combine the use of transcriptional and translational regulators to provide the same sort of multi-level control?

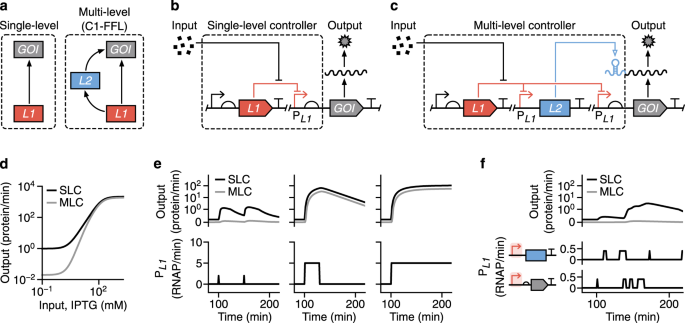
To test this idea, I realised that I’d need a way to build many different designs to figure out what combination of regulators worked best. Specifically, I chose to create designs that combined an inducible promoter, whose transcriptional activity is regulated by a repressor protein that is sensitive to the inducer, with an additional RNA-based regulator to create what we termed a ‘Multi-Level Controller’ (MLC). Two identical inducible promoters were used to express the mRNA of a gene of interest (GOI) and the RNA-based controller, in the form of a small RNA, which could then activate translation of the mRNA. As a result, the gene (we chose every synthetic biologist’s favourite, gfp) can only be expressed when both the inducible promoters are active and sufficient RNA regulator has been produced.
The two inducible promoters in our design are activated by the same input. Why would this be better than simply controlling the output gene with a single inducible promoter? The reason is that our more complex regulatory system implements what mathematicians call a coherent type-1 feedforward loop – a motif common in natural gene regulatory networks that is known to enable stringent control. For example, transcriptional noise could result in the accidental activation of a promoter. However, because the chance of this random event happening simultaneously for both promoters is tiny, the gene stays off – just like with the trigger hairs of the Venus flytrap!

The mechanism of RNA-based controllers we used is based on the formation of secondary structures of either RNA or DNA upstream of a target gene, which are altered when small triggering or activating RNAs are present at a sufficient concentration. Depending on the type of regulator, the secondary structures impede transcription, translation or both. In our case, our MLCs used either a toehold switch (THS) (Green et al. Cell 159, 925-39, 2014), small transcription activating RNAs (STAR) (Chappell et al. Nature Chemical Biology 11, 214-20, 2015), or what was called a dual control (DC) system (Westbrook et al. Nucleic Acids Research 45, 5614-24, 2017), which controlled both transcription and translation.
To make it easier for other researchers to build their own MLCs, I created a flexible design that was versatile and easy to build and screen. This consisted of an 8-part genetic template and a toolkit of parts that could be rapidly pieced together by Golden Gate assembly. The modularity of this toolkit means that you can create a wide range of complex constructs according to your preferences. The backbone without a controller present also constitutively expressed an orange fluorescent protein that drops out when the assembly is successful to make it easier to find correctly assembled designs.

Using this approach, I systematically built and tested a variety of MLCs that used different types of transcriptional and translational control. As a reference, I also included our original inducible system that simply used a repressor protein to control transcription of the output gfp gene.
What we found was that by regulating gene expression on two different levels, a far more stringent control was achieved. We saw virtually no leaky expression when the system was ‘off’ and strong expression when it was ‘on’. Some of our designs were able to provide a >2000-fold change in expression, making them some of the most high-performance systems built to date. Furthermore, when we characterised how the MLC systems responded to varying levels of input (i.e. inducer concentrations), we found that their response curves had a much sharper transition between ‘on’ and ‘off’ states. This digital-like response is a huge benefit when building genetic circuits that use digital (on/off) signals as they help ensure signals are properly propagated, reducing the chance of a malfunction.

But there’s more. We posted a preprint of these results on bioRxiv and were immediately contacted by Dr Amir Pandi, who was keen to try our MLCs in cell-free systems. Intrigued by his idea, we shipped over our genetic devices and a few weeks later learnt that they had worked. All of our MLC designs showed lower basal expression and improved fold-changes compared to an inducible promoter on its own – further highlighting the possibility to use these devices in many different contexts.

This project started with a problem – a misbehaving inducible system. By taking inspiration from nature, we were able to solve this and gain some added benefits. So, if you are looking for a new starting point for investigating the synergies that different gene expression processes and parts bring when combined, our paper is for you!
Further information
F. Veronica Greco, Amir Pandi, Tobias J. Erb, Claire S. Grierson, Thomas Gorochowski. Nature Communications. (DOI: 10.1038/s41467-021-21995-7)
Biocompute Lab: http://www.biocomputelab.org






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in