In the article presented in Communications Biology (DOI: 10.1038/s42003-020-01172-0), we report the whole genome information of Japanese muskmelon ‘Earl’s favorite Harukei-3’. This melon originates from a line that was imported from United Kingdom around 100 years ago. Although the original melon line was absent or not commercially used any more in UK, many high-grade muskmelon cultivars or varieties in Japan are developed based on it. If grown in an appropriate season, such Earl’s favorite series muskmelons or its derivatives produce fruits with very beautiful netted-appearance and sweet taste that are clearly different from other melons. Among the Earl’s favorite series muskmelons, Harukei-3 is an especially important line as it has been utilized as a standard line for breeding of high-grade muskmelon. Indeed, some high-grade muskmelons can be sold as a gift at very expensive prices (e.g. 200 USD for one fruit). Many popular consumer melons are also developed based on the Earl’s favorite series.
Over the last few years, DNA sequencing technology has been rapidly developed. Since its appearance of single molecule sequencing technology such as Oxford Nanopore Technology (ONT) and Pacbio, genome assembly study of eukaryotic organisms including higher plants has become much easier than before as they can sequence long DNA molecules. In particular, appearance of ONT Minion device was a sensational as it is a handy-type DNA sequencing instrument whose price is around 1,000 US dollar. Coupled with flow cells, this device can perform sequencing of both genomic DNA and RNA, which enables genomics study without purchasing expensive instruments. In case of genome DNA sequencing, ONT can obtain reads with N50 value > 10 kb if long and intact genomic DNA molecules are successfully purified and used as an input. Lengths of some reads exceed even 100 kb. Although ONT data are known to contain much higher frequency of sequence errors compared with Illumina short reads and Pacbio long reads, recent development of error correcting software and/or newer R10.x flow cells overcome such disadvantage to some degrees.
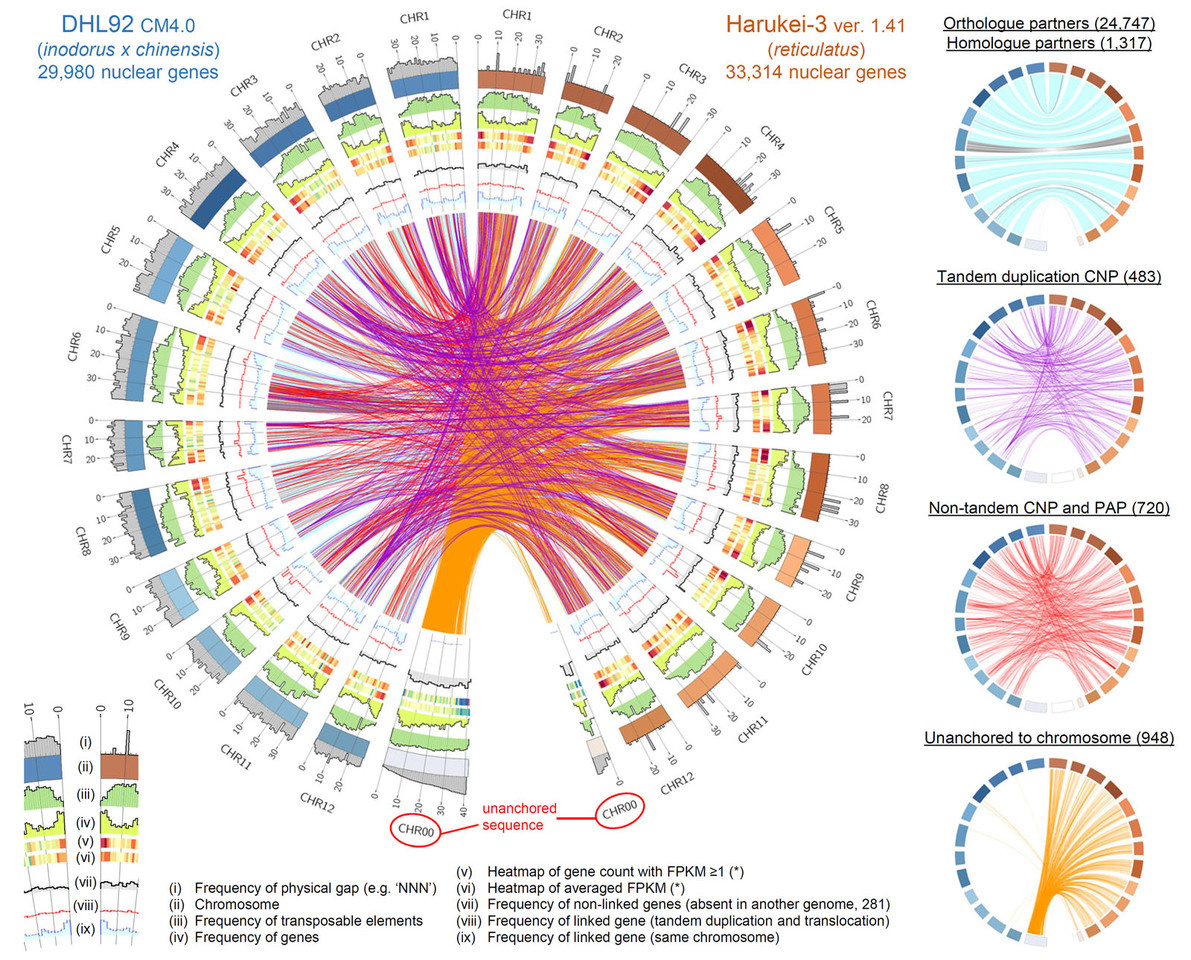
By using ONT for assembly of Harukei-3 melon, we assembled the 12 chromosomes of this melon genome with only 94 gaps (378 Mb in total). This high-grade genome assembly enabled construction of high-resolution tissue-wide RNA-seq atlas as well as detailed comparison with the previously published melon genomes. In our study, ONT also enabled contig assembly in additional seven melon accessions. Surprisingly, only one or two Minion flow cells were required for the analysis of each melon (in total nine flow cells used) although N50 varied 3.9 to 10.2 Mb. Our study demonstrated that these assemblies were enough to resolve structural genomic variation such as presence/absence polymorphisms (PAPs) (Figure 1). Indeed, we found genome-wide PAPs of retrotransposon sequences among melon accessions. Tissue-wide RNA-seq atlas showed that significant portion of such retrotransposon sequences were specifically transcribed in ripening fruits, with 59.4% of the neighboring genes showing similar expression patterns (r > 0.8). Taken together, we suggested that retrotransposons contributed to the modification of gene expression during diversification of melon genomes. It may also affect fruit ripening-inducible gene expression.

Figure 1. Presence/absence polymorphisms of retrotransposon sequences among melon accessions.
Undoubtedly, ONT opens the era of ‘personal’ genomics study. From DNA isolation to library preparation, it takes only 2-3 hours in a standard protocol. Recently, in tomato, sequencing of 100 genomes was reported by using ONT Promethion platform, which leads to the identification of structural variations beyond single nucleotide polymorphisms1. Indeed, some of the structural variations were shown to associate with the regulation of well-known QTL gene. In human, a telomere-to-telomere assembly of complete X chromosome was reported using ONT2. It is likely that more detailed mechanisms of genome evolution and diversification can be uncovered by ONT together with other technologies.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in