Over a few decades, the pioneering efforts of Medawar, Murray, Calne, Morris, Hamburger, Starzl, Barnard, Najarian, Terasaki, Stiller and numerous others have propelled transplantation from cautious experiment to the treatment of choice for vital organ failure1. Many seriously ill patients now enjoy superb immediate success, returning to virtually normal health with survival, quality of life and economic costs superior to those offered by any alternative therapy2. However, despite excellent short-term outcomes, less than half of these patients are alive with a functioning graft more than a decade later3. The reasons are varied, but the principal cause of graft loss remains rejection, of which the most challenging form is antibody-mediated rejection (AMR)4.
The approach to solving complex biomedical problems by rigorous research has also changed profoundly during this period, evolving from a largely single-investigator model to integrated teams working collaboratively to address common goals. Genome Canada, led by its first Chairs, Henry Friesen and Calvin Stiller, has been a powerful advocate of this transition, supporting large-scale research integrating molecular, clinical, population and computational sciences. The Genome Canada Transplant Consortium (www.gctransplant.ca, @genomecanadatx), from which our current original article derives, is an example of the results from this model, drawing together over 70 principal investigators from 22 leading universities in Canada, the USA, EU and UK to work on the common theme of AMR. The program engages four pillars, namely structural biology, laboratory diagnostics, systems pharmacology, and social sciences research, exploring a combination of strategies designed to prevent the devastating complications of AMR in the kidney and, subsequently, in other transplanted organs.
The present article draws on the principles of structural biology to explore the targets of antibody binding, known as epitopes or “eplets”, and to determine whether achieving donor and recipient compatibility by matching at these structures is a practical reality (Figure 1). Like all important advances, it builds on the work of other leaders in the field including Rene Duquesnoy, Frans Claas, Steven Marsh and Derek Middleton who have worked tirelessly to define the structure-function relationships of HLA5–8, and of our fellow Canadian researchers Chris Wiebe, Peter Nickerson, Kathryn Tinckam, Ruth Sapir-Pichhadze and others who have laid the groundwork to explore the use of B-cell epitopes in graft survival9–11.

Figure 1. Structural biology analysis to identify HLA B cell epitopes. Modern omics technologies enable the linkage between genomics, proteomics and function related to tertiary structure. In our Communications Biology paper, we utilized high-throughput sequencing data of the HLA genes to identify essential structural determinants in antibody binding. Nucleotide sequences were converted into amino acids and B cell epitopes were identified. These determinants were identified on 3D protein structures retrieved from the Protein Databank to elucidate the targets of antibodies in transplant rejection.
The article describes the important reduction in complexity as we move from HLA alleles, of which over 25,000 are now recognized, to epitopes, of which approximately 150 have been verified as antibody targets (Figure 2). It demonstrates substantial sharing of these epitopes between products of the HLA Class I genes (HLA-A, B and C), and more limited sharing within certain HLA Class II genes (HLA-DRB1/3/4/5, DQA1, DQB1, DPA1, and DPB1). The models employed show that current donor-recipient organ sharing programs, which do not prioritize HLA compatibility due to the difficulty in achieving matching across the large number of HLA alleles, may expect very low epitope compatibility leading to sub-optimal clinical outcomes. However, they confirm the exciting potential that high degrees of compatibility are achievable by deliberately matching for these epitopes. This is particularly true for the Class II HLA-DR and DQ alleles which Wiebe, Nickerson and colleagues have shown to be cardinal in graft success12.

Figure 2. The reduction in HLA complexity when alleles are converted into epitopes. Upper portions of each Circos diagram show the HLA alleles identified in the study transplant population (n=1846), while the lower portions show the translation into the 150 antibody-verified epitopes, simplifying the process for prospective patient and donor matching.
And perhaps most importantly, the models indicate that high-level compatibility, including a zero epitope/eplet mismatch at the HLA-DR and DQ loci, can be achieved within the waiting list size of most major transplant programs around the world. This mitigates the requirement for routine sharing of organs on a national or international basis with the attendant challenges of logistics, costs, and graft deterioration. Technological advances in PCR and new Nanopore typing13 methods now in clinical translation promise high-resolution HLA typing of deceased donors within 6 hours, a critical component of epitope-based matching. And the implementation of web-based tools by Canadian Blood Services will provide the operational support for epitope-based donor allocation to all sites across the country.
But improved matching is simply the first important step. The pace of biomarker development to predict or detect the immune response against donor antigens has accelerated around the world, and innovations such as T-cell receptor sequencing, solid-phase antibody monitoring, and donor-derived cell-free DNA detection may soon provide the precision to identify early rejection and to enable rapid and effective intervention. And systems pharmacology, well established within the field of oncology, offers the opportunity to integrate pharmacogenomics, kinetics, and dynamics to define personal needs and adjust care in a way not previously possible. If these innovations are successful in preventing the destructive process of graft rejection, opportunities arise for novel strategies to induce operational tolerance and eventually to avoid the ravages of long-term immune suppression.
The importance of this Genome Canada Consortium may extend far beyond the realm of transplantation, however. The immune system is a complex biological network that is vital to human health from embryogenesis to tissue repair and is key to our defense against major diseases including infection and cancer. Innate and adaptive immunity integrate seamlessly to ensure tolerance to our own cells and tissues, while quickly detecting and eliminating foreign chemicals, pathogens, or malignant cells that risk our health and survival. However, our complex immune system is not failsafe, and dysfunction manifest by changes in amplitude or specificity lead to a constellation of immune-related diseases that currently affect over 10% of Canada’s population with vast economic costs14.
The lightning pace of research in genomics, epigenetics, proteomics, and cell biology has driven our growing understanding of the enormous complexity of our immune system and spurred ground-breaking discoveries in the fields of stem cell transplantation, solid organ transplantation, autoimmune disease and inflammation, infectious immunity, immune-oncology, and immune deficiency disorders. The major advances from immune research in new biologics, vaccines, cellular therapeutics, and many other cutting-edge innovations provide a superb opportunity to develop integrated strategies in Precision Medicine and avert the life-threatening consequences of immune-mediated tissue injury.
And yet the domain of immunology remains highly fragmented and investigators in these fields continue to work in silos. New understandings, promising diagnostics and highly innovative biological therapies – single-cell omics, immune cell receptor sequencing, innovative biologics targeting novel cell subsets and cytokine pathways, CAR-T cells adapted to autoantibody receptors or regulation, and genomic base editing – remain largely confined to individual laboratories or disease areas. As a result, enormous opportunities for collaborative research to accelerate translation across multiple fields are missed.
The field of cancer medicine has led a revolution in Precision Medicine, linking research and care in complex multifaceted diseases through the integration of basic science, translational research, and care delivery within a single operational framework. Canada’s Cancer Centres have embraced this concept and are now among the leading research and care facilities in the world. Our current Genome Canada program has successfully adopted this strategy in the focused field of kidney replacement and we are now working to extend this approach to a novel International Network in Immune Sciences. This effort will link leading universities across North America, Europe, and emerging nations to apply Precision Medicine strategies across the broader fields of autoimmunity, infectious immunity, transplant immunity, and immuno-oncology (Figure 3). Moreover, it will provide a model to ensure the integration and clinical application of research in these fields, along with a detailed and continuous evaluation of the societal costs and benefits.

Figure 3. The principle domains of immune-related disease targeted by the planned International Network in Immune Sciences. This program will link investigators, healthcare providers, patients and policy makers across North America, Europe, and other nations in the fields of autoimmunity, transplant, tumor, and infectious immunity in a collaborative effort to coordinate research and care in these disorders.
References
- Barker, C. F. & Markmann, J. F. Historical overview of transplantation. Cold Spring Harb. Perspect. Med. 3, 1–18 (2013).
- Levy, A. R. et al. Projecting long-term graft and patient survival after transplantation. Value Heal. 17, 254–260 (2014).
- Organ replacement in Canada: CORR annual statistics, 2019 | CIHI. https://www.cihi.ca/en/organ-replacement-in-canada-corr-annual-statistics-2019.
- Sellarés, J. et al. Understanding the causes of kidney transplant failure: The dominant role of antibody-mediated rejection and nonadherence. Am. J. Transplant. 12, 388–399 (2012).
- Duquesnoy, R. J. HLA epitope based matching for transplantation. Transpl. Immunol. 31, 1–6 (2014).
- Kramer, C. S. M. et al. The long and winding road towards epitope matching in clinical transplantation. Transpl. Int. 32, 16–24 (2019).
- Robinson, J. et al. IPD-IMGT/HLA Database. Nucleic Acids Res. 48, D948–D955 (2020).
- Gonzalez-Galarza, F. F., Christmas, S., Middleton, D. & Jones, A. R. Allele frequency net: A database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 39, 913–919 (2011).
- Wiebe, C. et al. Evidence for the alloimmune basis and prognostic significance of Borderline T cell–mediated rejection. Am. J. Transplant. 2499–2508 (2020) doi:10.1111/ajt.15860.
- Sapir-Pichhadze, R. et al. Epitopes as characterized by antibody-verified eplet mismatches determine risk of kidney transplant loss. Kidney Int. 97, 778–785 (2020).
- Sypek, M., Kausman, J., Holt, S. & Hughes, P. HLA Epitope Matching in Kidney Transplantation: An Overview for the General Nephrologist. American Journal of Kidney Diseases (2018) doi:10.1053/j.ajkd.2017.09.021.
- Wiebe, C. et al. HLA-DR/DQ molecular mismatch: A prognostic biomarker for primary alloimmunity. Am. J. Transplant. 19, 1708–1719 (2019).
- De Santis, D., Truong, L., Martinez, P. & D’Orsogna, L. Rapid high-resolution HLA genotyping by MinION Oxford nanopore sequencing for deceased donor organ allocation. Hla 96, 141–162 (2020).
- Jacobs, P., Bissonnette, R. & Guenther, L. C. Socioeconomic burden of immune-mediated inflammatory diseases - Focusing on work productivity and disability. J. Rheumatol. 38, 55–61 (2011).
Acknowledgements
We are indebted to the members of the Genome Canada Transplant Consortium for their contribution to this research. Research was supported by Genome Canada, Genome British Columbia, Genome Quebec, and Genome Alberta, Canadian Institutes of Health Research, and partnered grants from Omixon.
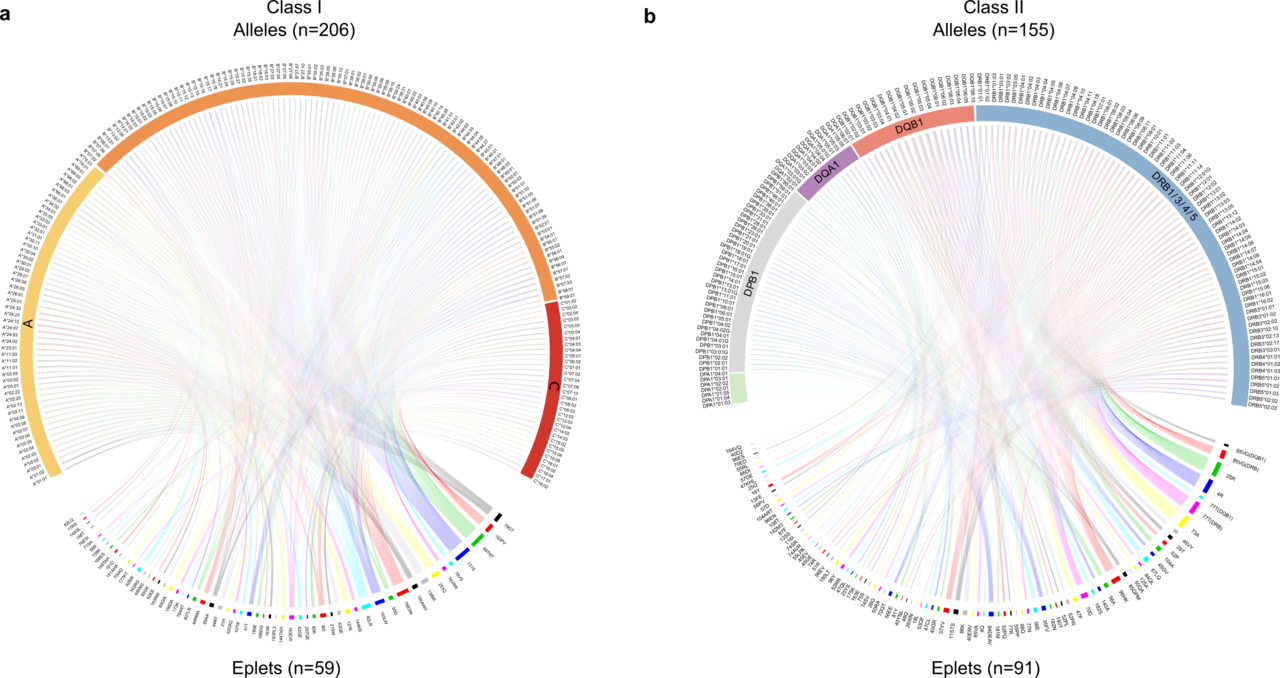







Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in