It’s the capacity of pluripotent stem cells to generate any cell in the adult body that makes stem cell biology such a fascinating and promising field. By thoughtful construction of efficient, controlled, developmentally directed differentiations, scientists have been able to guide pluripotent cells toward many desired somatic cell types; thereby opening up possibilities for disease modeling, drug screening, cell delivery, and developmental studies. However, these opportunities are only possible if there exist reliable differentiation protocols to lineages of interest. Within the field of stem cell biology, a few lineages remain profoundly understudied; tendon is one of them.
Our laboratory’s research focuses on the field of tendon regeneration. Though tendons are critical components of the musculoskeletal system they are consistently overlooked. Large scale transcriptomic profiling efforts such as ENCODE and single-cell RNA sequencing atlases consistently omit dense connective tissues such as tendon and fibrocartilage (Han et al., 2018; Tabula Muris et al., 2018; Yue et al., 2014). As a result, the master regulators that govern the tendon fate haven’t been identified and many of the molecular events and signaling pathways that specify tendon induction are unknown.
As tendon biologists, we are excited by the prospect of modeling tendinopathies, screening pro-regenerative drugs, and designing therapeutic cell delivery strategies for acute tendon injuries. However, this requires a differentiation protocol for a lineage whose developmental trajectory has many missing links. We were therefore posed with a challenging problem. To strategize around this, we utilized a two-step process for developing a tendon differentiation. First, we began by gathering insights from the available tendon development literature. Then, we used single-cell RNA sequencing to identify missing or uncontrolled signaling pathways that we could manipulate to improve efficiency and specificity. Finally, with a reliable and efficient differentiation, bulk RNA sequencing allowed for step-wise molecular profiling of fate specification; giving us desperately needed insight into the molecular control of fate induction.
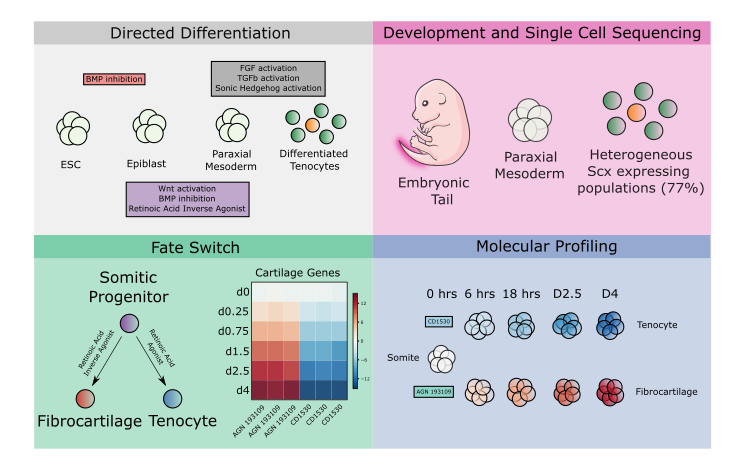
The basic framework for our initial differentiation protocol toward axial tendon came from research by Ronen Schweitzer’s lab which established a requirement for TGFb signaling in tendon induction (Pryce et al., 2009). We combined it with information surrounding somitic mesoderm induction (a progenitor for axial tendons) (Zhao et al., 2014). We chose to use single-cell sequencing technologies on both the product of our tendon differentiation and the E14.5 mouse tail. The developing tail tendons and their neighboring populations would serve as controls for transcriptionally profiling our cells and understanding how our populations related to one another. To our surprise, the sequencing showed a mixed population of tendon and fibrocartilage. Fibrocartilage is another cell type that gets omitted from sequencing atlases and very few efforts have been made to explore the tissue with next-generation sequencing technologies. Single-cell sequencing tendon and fibrocartilage in vivo and in vitro revealed differentially expressed genes that enrichment software predicted the retinoic acid pathway might control. Excitingly, we were able to control this fate switch use small molecule activators and inverse agonists of this pathway. To improve the efficiency of our differentiation we used our single-cell sequencing on the somite to examine heterogeneity. We believed that any heterogeneity in the somite might reveal a lack of control in the differentiation and an opportunity for improvement. Enrichment analysis on differentially expressed genes from our somite clusters, again, identified retinoic acid as the most activated pathway. When we addressed this by adding a retinoic acid antagonist to our somite differentiation and we were ultimately able to generate Scx expressing tendon and fibrocartilage with 97% efficiency.
Having optimized our differentiations, we set out to use this tool to answer some fundamental questions about tendon and fibrocartilage specification. We designed a bulk sequencing experiment to probe the fate trajectory of both our tendon and fibrocartilage conditions by taking incremental timepoints in both conditions starting at somite following through to our final differentiated state. A combination of unbiased and biased analyses decomposed a number of genetic programs that contributed to the early induction of both cell types as well as modules that were responsible for fate divergence. We found known factors in tendon and fibrocartilage among our identified genes such as Scx and Sox9, but we also found several genes with no previously researched role in either cell fate.

novel fates switch discovery, and molecular profiling of divergent fate induction
These modules of novel tendon and fibrocartilage genes open the door for numerous studies into the molecular underpinnings of the tendon fate. It is our hope by continuing to research the roles of these determined factors we may steadily fill in the story of what makes a tendon cell a tendon cell and develop strategies for tendon regeneration. As is the case with many other differentiation protocols, we hope that our 3-dimensional tensioned constructs can serve as a tool for understanding development and maturation from a biophysical perspective as well. We believe that our differentiation protocol will be a critical tool in the future of tendon research and we are excited to continue to probe at the many unknowns of tendon development.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in